 Economic principles in cell metabolism
Economic principles in cell metabolism
Metabolic value theory - a theory of optimal metabolic states
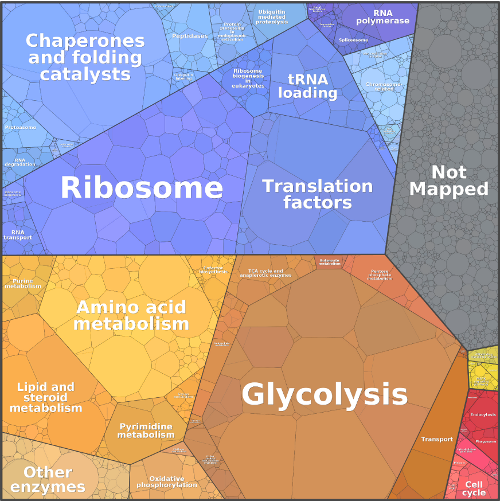
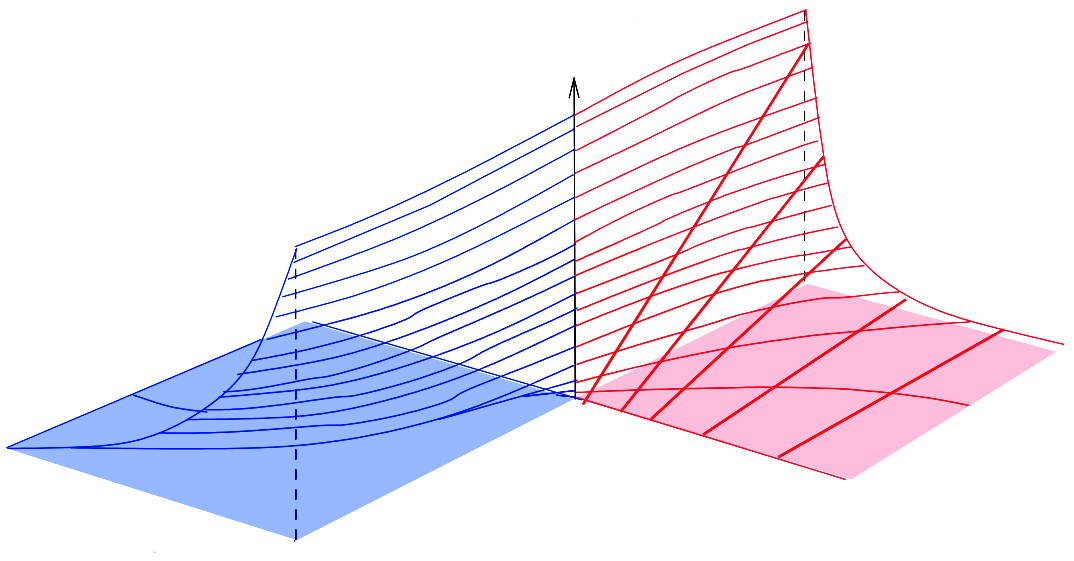
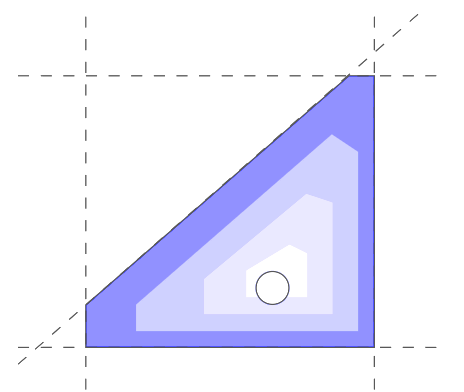
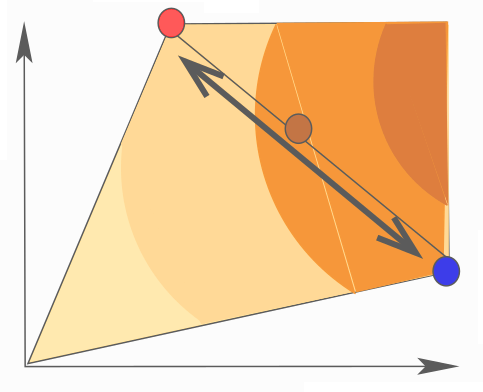
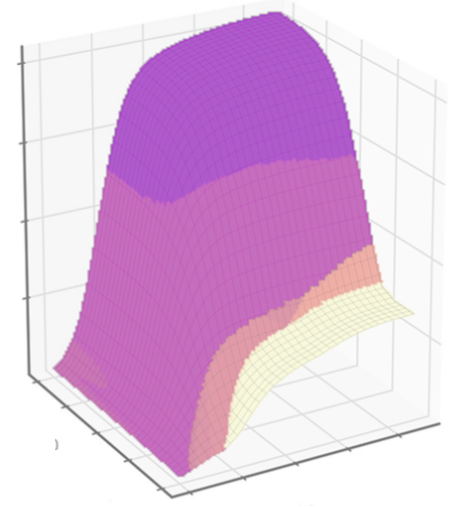
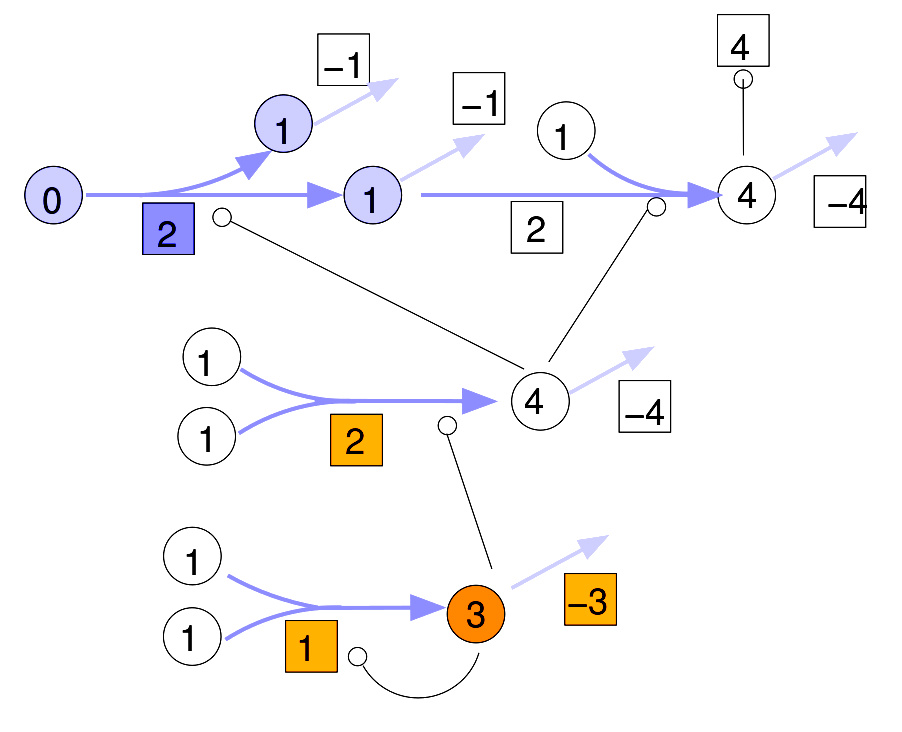
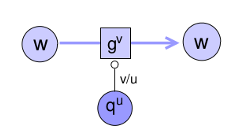
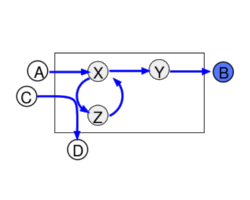
Metabolic systems are governed by compromises between metabolic benefit and enzyme cost. This hypothesis and its consequences can be studied by metabolic models in which enzyme profiles follow optimality principles. In optimal metabolic states, active enzymes must provide a benefit: higher enzyme levels must improve the metabolic objective to justify the additional enzyme cost. This entails general relations between metabolic fluxes, reaction elasticities, and enzyme costs. Similar laws hold for flux analysis and models of growing cells. The articles and preprints on this page adopt this principle and describe cellular resource allocation from different perspectives.
 |
 |
 |
 |
 |
|
 |
 |
 |
 |
 |
|
Read more:
- Poster
"Enzyme economy in metabolic networks" - Software Matlab code / Documentation
Wolfram Liebermeister (2023)