Structural thermokinetic modelling for metabolic models
Main | Models | Matlab code | Article
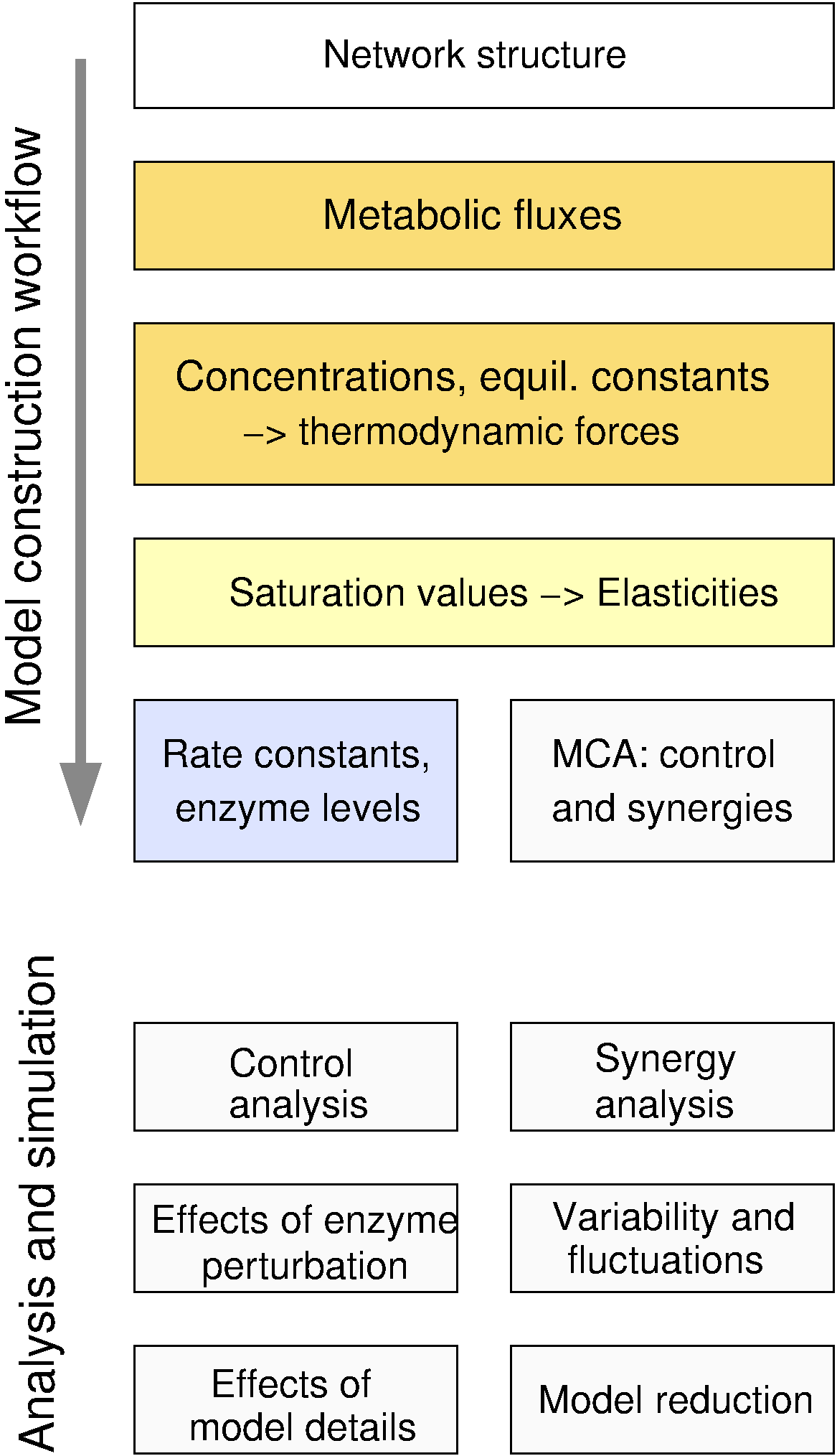 |
What is structural thermokinetic modelling?Metabolic networks can be turned into kinetic models in a predefined steady state by sampling the reaction elasticities in this state. Elasticities for many reversible rate laws can be computed from the reaction Gibbs free energies, which are determined by the state, and from physically unconstrained saturation values. Starting from a network structure with allosteric regulation and consistent metabolic fluxes and concentrations, and following the Structural Kinetic Modelling methodology (Steuer et al. 2006), one may sample the elasticities, compute the control coefficients, and reconstruct a kinetic model with consistent reversible rate laws. Some of the model variables are manually chosen, fitted to data, or optimised, while the others are computed from them. What can structural thermokinetic modelling be used for?The resulting model ensemble allows for probabilistic predictions, for instance, about possible dynamic behaviour. By adding more data or tighter constraints, the predictions can be made more precise. Model variants differing in network structure, flux distributions, thermodynamic forces, regulation, or rate laws can be realised by different model ensembles and compared by significance tests. The thermodynamic forces have specific effects on flux control, on the synergisms between enzymes, and on the emergence and propagation of metabolite fluctuations. Large kinetic models could help to simulate global metabolic dynamics and to predict the effects of enzyme inhibition, differential expression, genetic modifications, and their combinations on metabolic fluxes. |
Reference
If you use structural thermokinetic modelling in your work, please cite my article:
Liebermeister W. (2022), Structural thermokinetic modelling, Metabolites 12(5), 434.
Contact
Please contact Wolfram Liebermeister with any questions or comments.